
Next: GUI mode - spins Up: Setup in the GUI Previous: GUI mode - setting Contents Index
For this section, the example of protein 15N relaxation data will be used to illustrate how to set up the data structures.
To manipulate the molecule, residue and spin data structures in the GUI, the most convenient option is to use the spin viewer window (see Figure 1.10 on page ![[*]](crossref.png) ).
This window can be opened in four ways:
).
This window can be opened in four ways:
You will then see:
|
At this point, click on the “Load spins” button (or the “Load spins” menu entry from the right click pop up menu) to launch the spin loading wizard. A number of options will be presented to you:
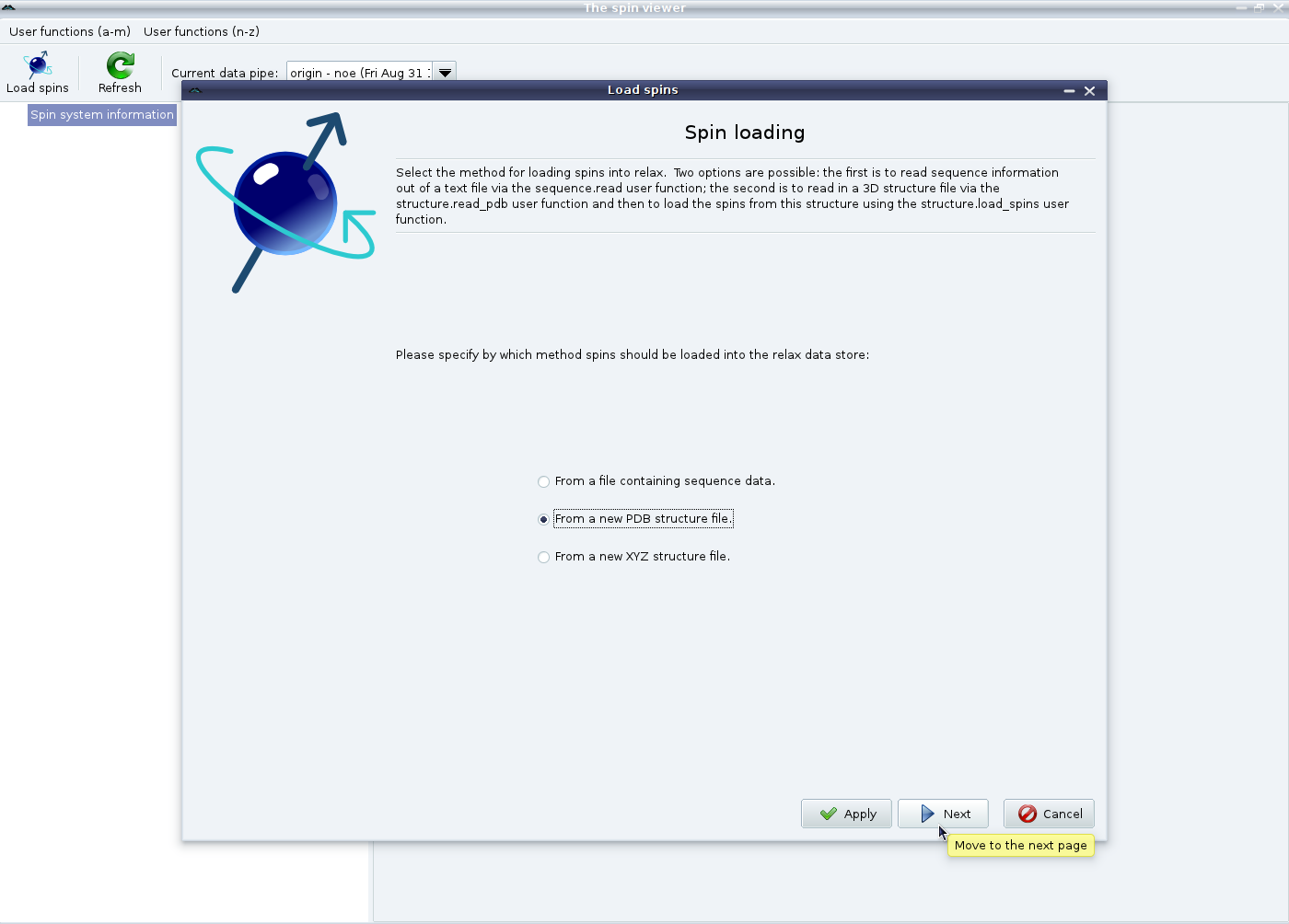
|
Here the spins will be loaded from a PDB file. If you do not have a 3D structure file, please see the next section. After selecting “From a new PDB structure file” and clicking on “Next”, you will see:
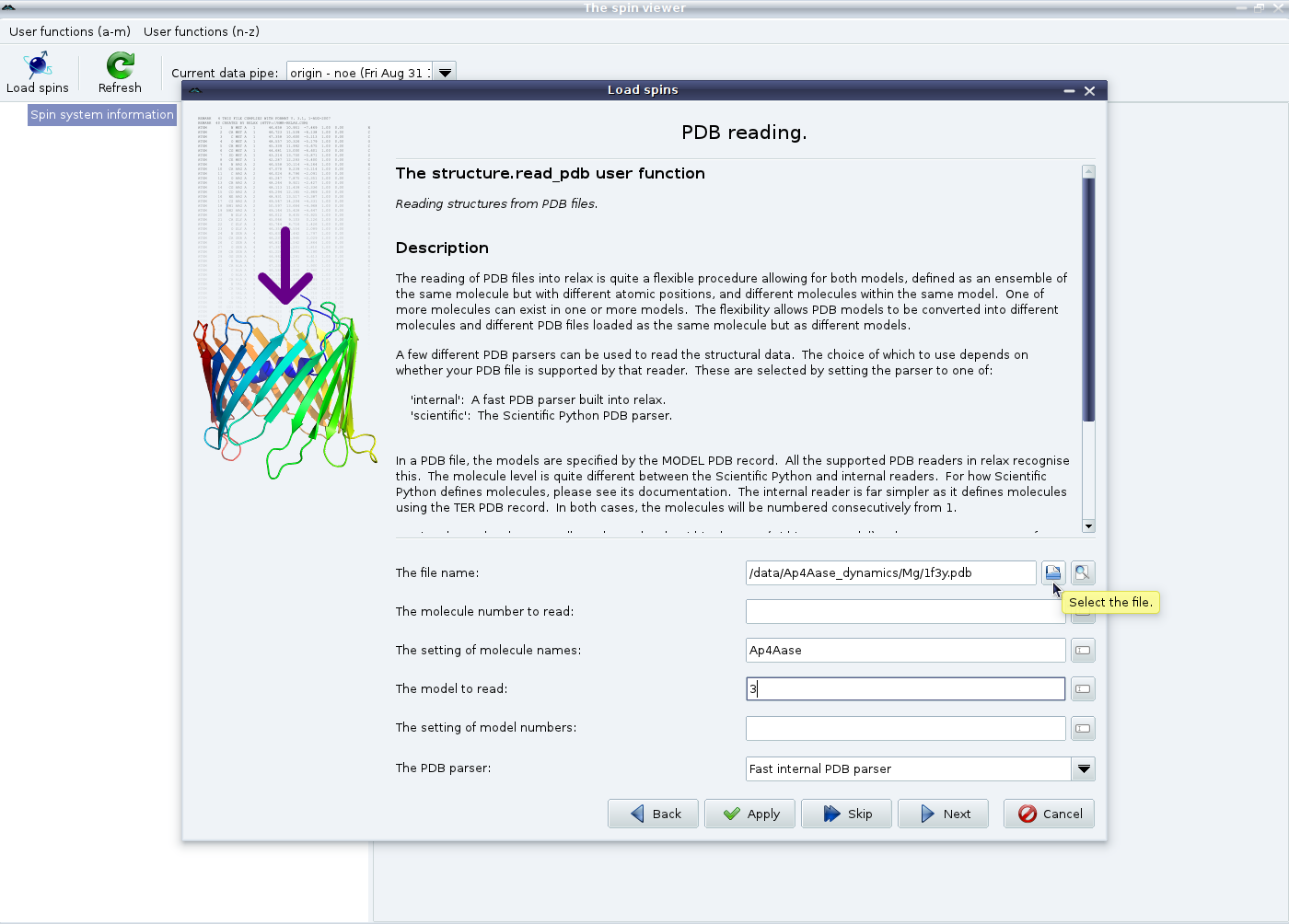
|
Now select the PDB file you wish to use. The other options in this screen allow you to handle NMR models and multiple molecules within a single PDB file. These options are explained in the window. Hovering the mouse over the options will give additional hints. In this example, the 3 model from the 1F3Y PDB file will be read and the single molecule will be named “Ap4Aase” to override the default naming of “1f3y_mol1”. Now click on “Next” to bring up the spin loading page:
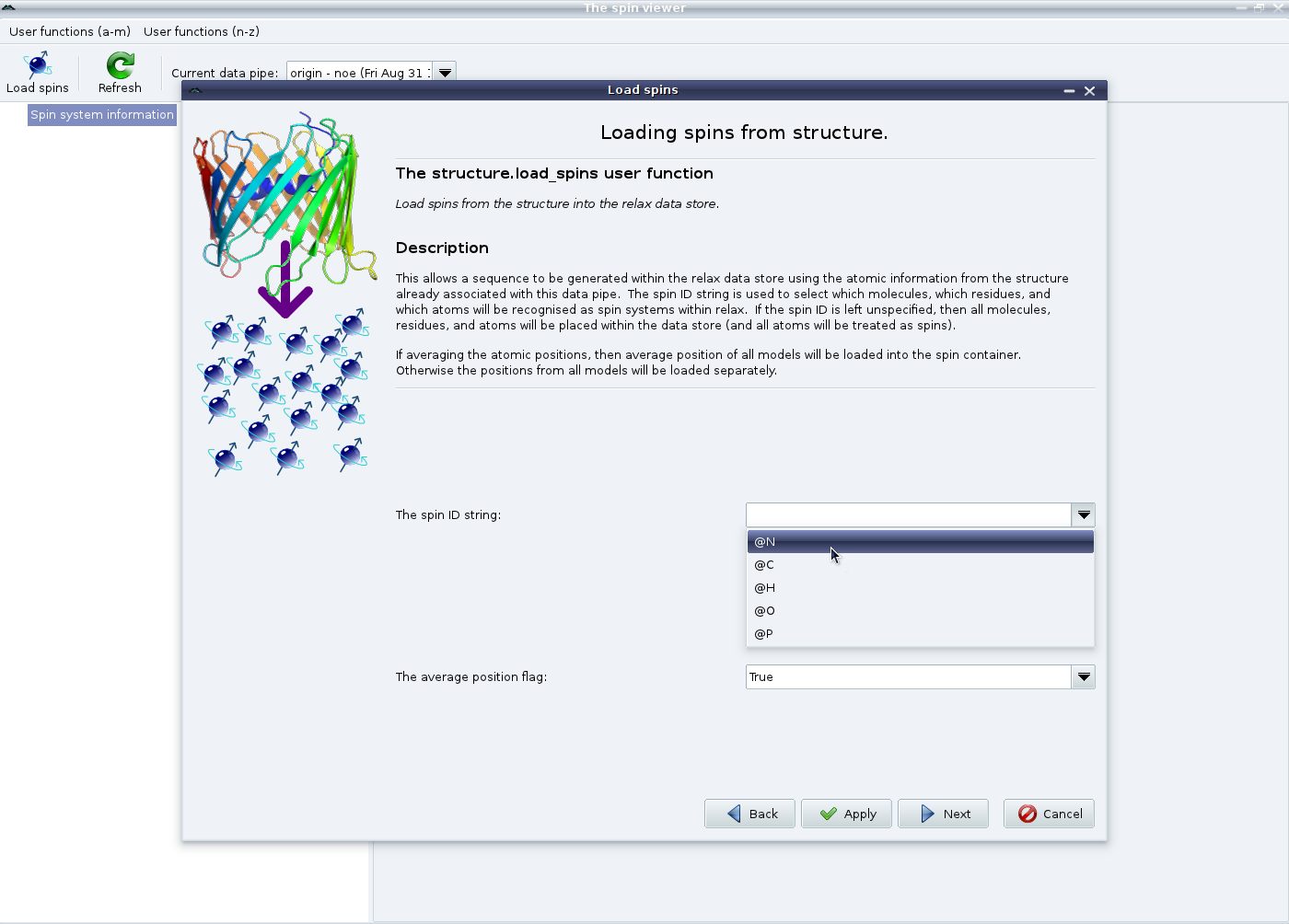
|
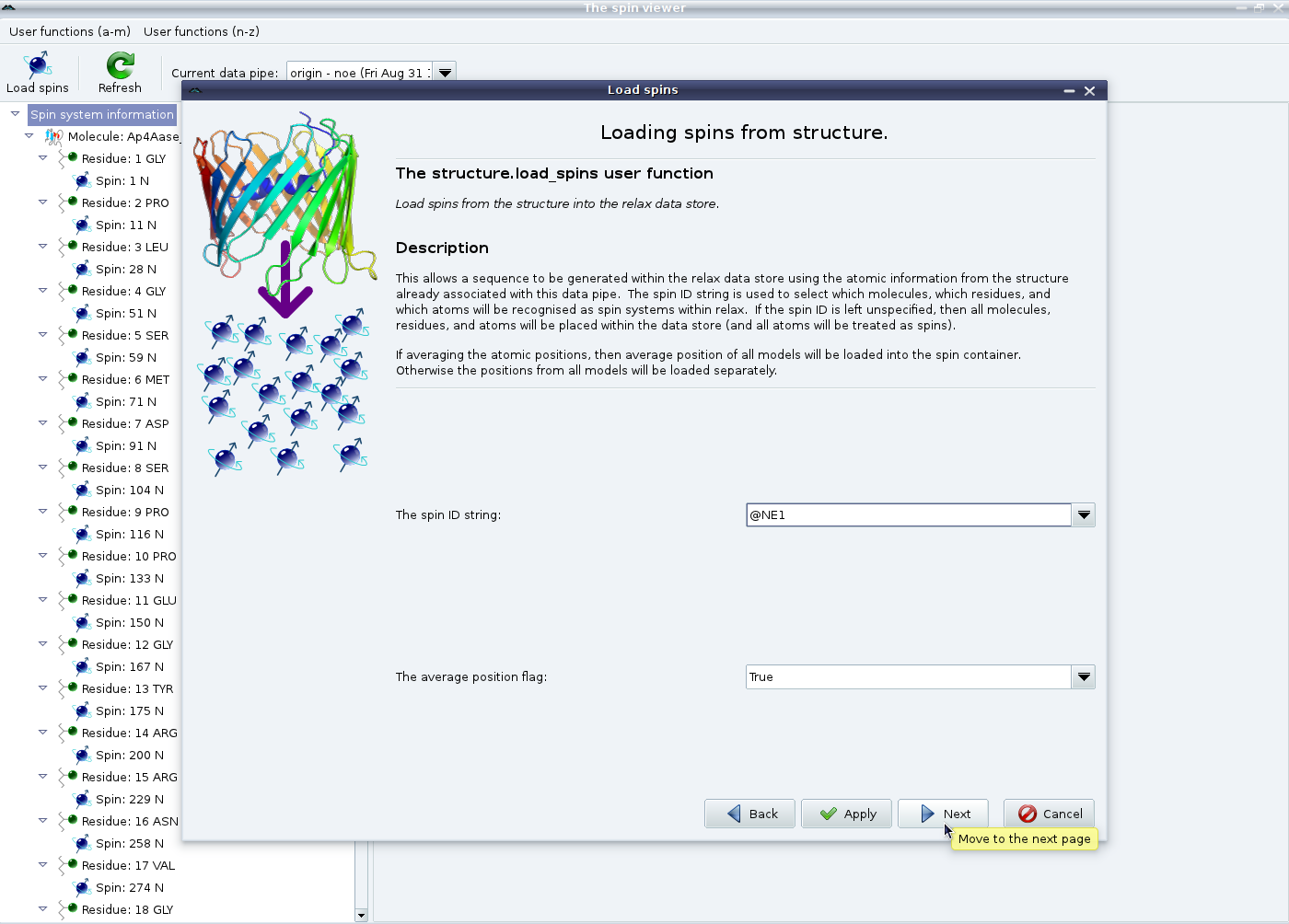
This is a bit more complicated. In this example we are studying the backbone dynamics of 15N spins of a protein. Therefore first set the spin ID string to “@N” (which can be selected from the pull down) and click on “Apply” to set up the backbone spins. Do not click on “Next” yet. If the current study requires the specification of the dipole-dipole interaction (for example if it involves relaxation data - model-free analyses, consistency testing, reduced spectral density mapping; or the dipolar coupling - the N-state model or ensemble analyses, the Frame Order theory) you will also need to load the 1H spins as well. Therefore set the spin ID string to “@H” and click on “Apply” again.
|
Now change the spin ID string to “@NE1” and then click on “Next” (or “Apply” if the Trp protons “@HE1” need to be loaded as well). This will add spin containers for the tryptophan indole 15N spins. Finally click on “Finish” to exit the wizard:
|
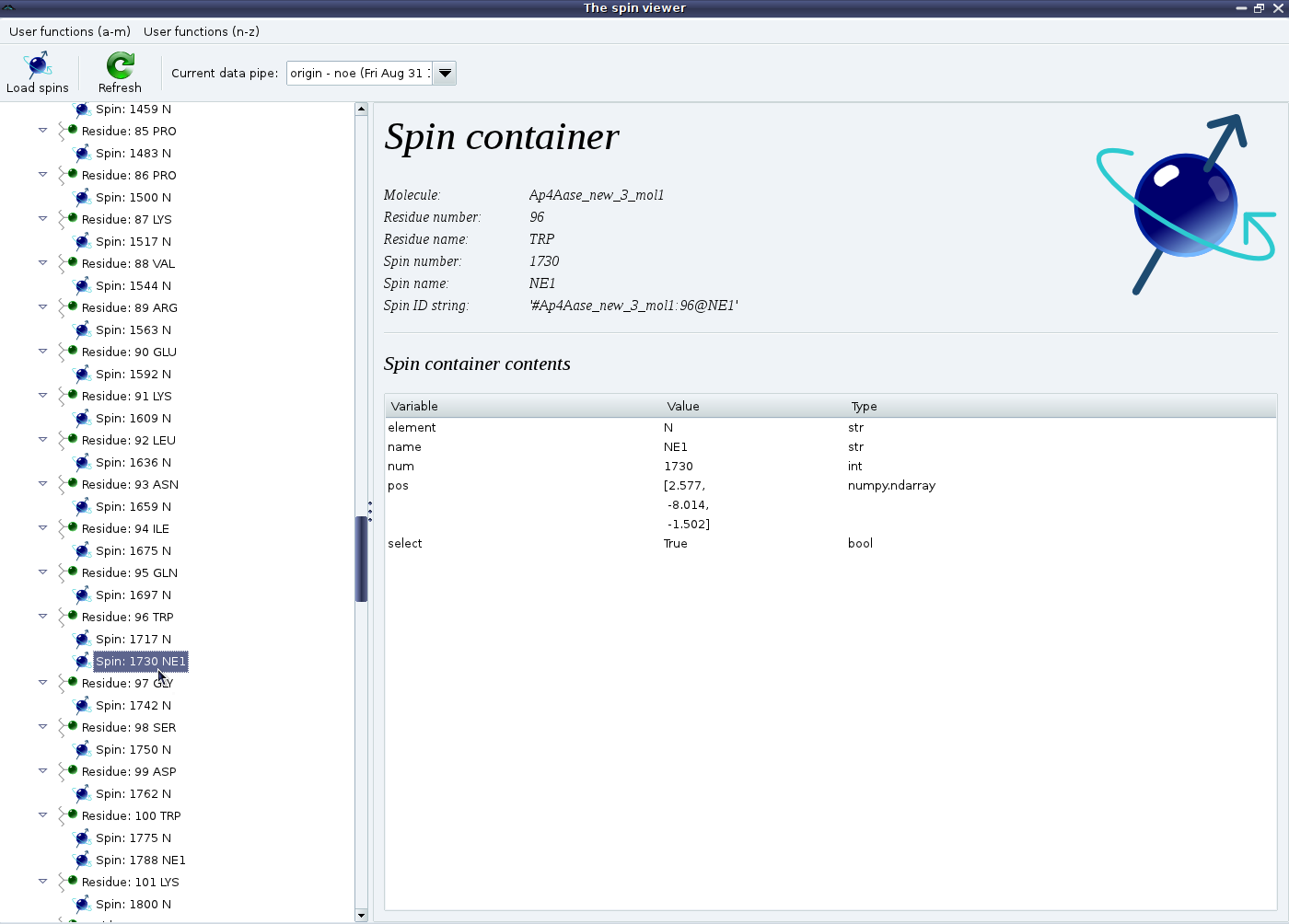
You should now see something such as:
|
If the 1H spins have been loaded as well, then you should see exactly twice as many spin containers as shown above.
The relax user manual (PDF), created 2024-06-08.