|
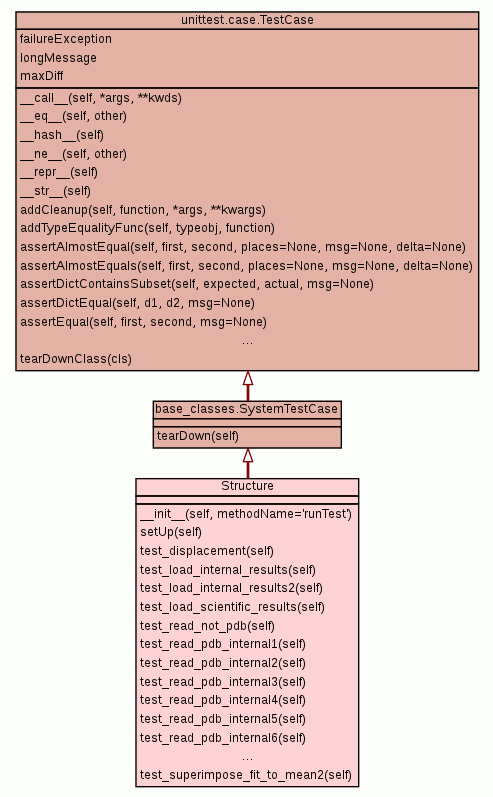
Inherited from base_classes.SystemTestCase:
tearDown
Inherited from unittest.case.TestCase:
__call__,
__eq__,
__hash__,
__ne__,
__repr__,
__str__,
addCleanup,
addTypeEqualityFunc,
assertAlmostEqual,
assertAlmostEquals,
assertDictContainsSubset,
assertDictEqual,
assertEqual,
assertEquals,
assertFalse,
assertGreater,
assertGreaterEqual,
assertIn,
assertIs,
assertIsInstance,
assertIsNone,
assertIsNot,
assertIsNotNone,
assertItemsEqual,
assertLess,
assertLessEqual,
assertListEqual,
assertMultiLineEqual,
assertNotAlmostEqual,
assertNotAlmostEquals,
assertNotEqual,
assertNotEquals,
assertNotIn,
assertNotIsInstance,
assertNotRegexpMatches,
assertRaises,
assertRaisesRegexp,
assertRegexpMatches,
assertSequenceEqual,
assertSetEqual,
assertTrue,
assertTupleEqual,
assert_,
countTestCases,
debug,
defaultTestResult,
doCleanups,
fail,
failIf,
failIfAlmostEqual,
failIfEqual,
failUnless,
failUnlessAlmostEqual,
failUnlessEqual,
failUnlessRaises,
id,
run,
shortDescription,
skipTest
Inherited from unittest.case.TestCase (private):
_addSkip,
_baseAssertEqual,
_deprecate,
_formatMessage,
_getAssertEqualityFunc,
_truncateMessage
Inherited from object:
__delattr__,
__format__,
__getattribute__,
__new__,
__reduce__,
__reduce_ex__,
__setattr__,
__sizeof__,
__subclasshook__
|