| Trees | Indices | Help |
|
|---|
|
|

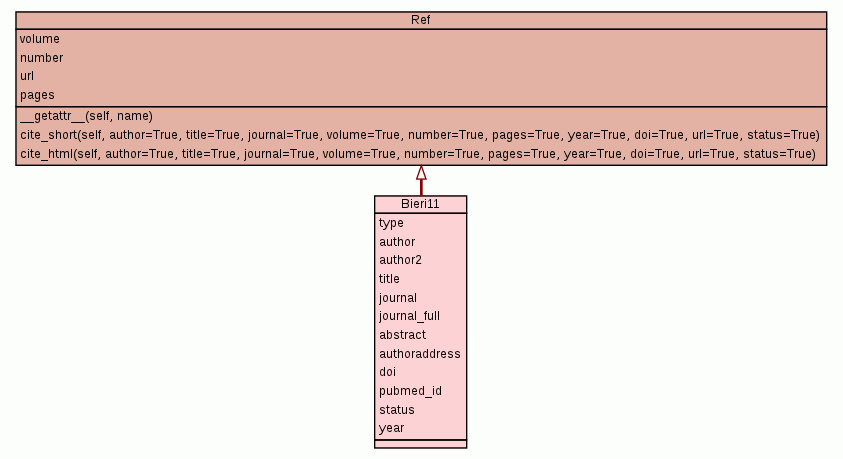
Bibliography container.
|
|||
|
Inherited from |
|||
|
|||
type = hash(x) |
|||
author = hash(x) |
|||
author2 = hash(x) |
|||
title = hash(x) |
|||
journal = hash(x) |
|||
journal_full = hash(x) |
|||
abstract = |
|||
authoraddress = |
|||
doi = hash(x) |
|||
pubmed_id = 21618018hash(x) |
|||
status = hash(x) |
|||
year = 2011hash(x) |
|||
|
|||
author2hash(x)
|
titlehash(x)
|
abstract
|
authoraddress
|
| Trees | Indices | Help |
|
|---|
| Generated by Epydoc 3.0.1 on Wed Apr 10 14:15:51 2013 | http://epydoc.sourceforge.net |