
Next: structure.add_helix Up: The list of functions Previous: statistics.model Contents Index
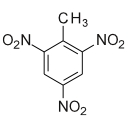

Add an atom.
mol_name: The name of molecule container to create or add the atom to.
atom_name: The atom name.
res_name: The residue name.
res_num: The residue number.
pos: The atomic coordinates. For specifying different coordinates for each model of the ensemble, a list of lists can be supplied.
element: The element name.
atom_num: The optional atom number.
chain_id: The optional chain ID string.
segment_id: The optional segment ID string.
pdb_record: The optional PDB record name, e.g. `ATOM' or `HETATM'.
This allows atoms to be added to the internal structural object. To use the same atomic coordinates for all models, the atomic position can be an array of 3 values. Alternatively different coordinates can be used for each model if the atomic position is a rank-2 array where the first dimension matches the number of models currently present.