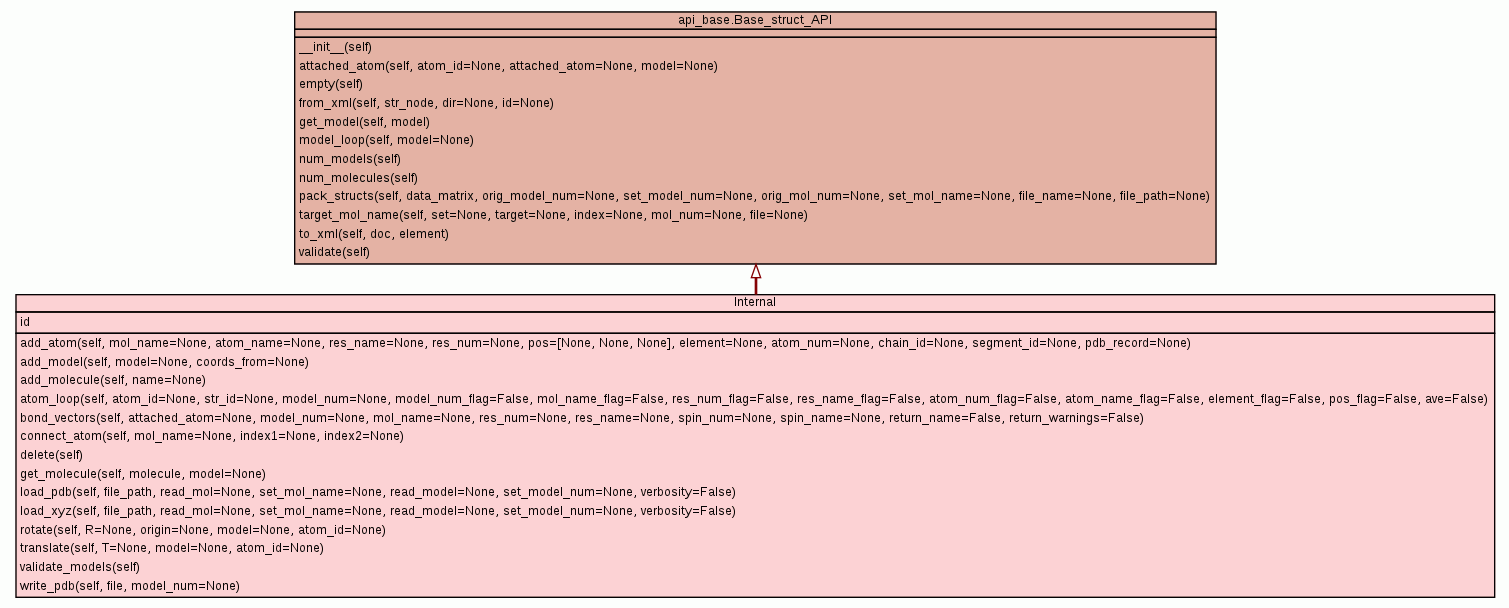
The internal relax structural data object.
The structural data object for this class is a container possessing a
number of different arrays corresponding to different structural
information. These objects are described in the structural container
docstring.
|
tuple consisting of the atom number (int), atom name (str), element
name (str), and atomic position (Numeric array of len 3)
|
_bonded_atom(self,
attached_atom,
index,
mol)
Find the atom named attached_atom directly bonded to the atom located
at the index. |
source code
|
|
|
|
|
|
str or None
|
|
|
tuple of int and array of str
|
|
|
tuple of int and array of str
|
|
|
tuple of int and list of str
|
_parse_mols(self,
records)
Generator function for looping over the molecules in the PDB records
of a model. |
source code
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
str
|
|
|
|
add_atom(self,
mol_name=None,
atom_name=None,
res_name=None,
res_num=None,
pos=[None, None, None],
element=None,
atom_num=None,
chain_id=None,
segment_id=None,
pdb_record=None)
Add a new atom to the structural data object. |
source code
|
|
|
ModelContainer instance
|
|
|
|
|
|
tuple consisting of optional molecule name (str), residue number
(int), residue name (str), atom number (int), atom name(str), element
name (str), and atomic position (array of len 3).
|
atom_loop(self,
atom_id=None,
str_id=None,
model_num=None,
model_num_flag=False,
mol_name_flag=False,
res_num_flag=False,
res_name_flag=False,
atom_num_flag=False,
atom_name_flag=False,
element_flag=False,
pos_flag=False,
ave=False)
Generator function for looping over all atoms in the internal relax
structural object. |
source code
|
|
|
list of numpy arrays (or a tuple if return_name or return_warnings
are set)
|
bond_vectors(self,
attached_atom=None,
model_num=None,
mol_name=None,
res_num=None,
res_name=None,
spin_num=None,
spin_name=None,
return_name=False,
return_warnings=False)
Find the bond vector between the atoms of 'attached_atom' and
'atom_id'. |
source code
|
|
|
|
connect_atom(self,
mol_name=None,
index1=None,
index2=None)
Connect two atoms in the structural data object. |
source code
|
|
|
|
|
|
MolContainer instance or None
|
|
|
bool
|
load_pdb(self,
file_path,
read_mol=None,
set_mol_name=None,
read_model=None,
set_model_num=None,
verbosity=False)
Method for loading structures from a PDB file. |
source code
|
|
|
bool
|
load_xyz(self,
file_path,
read_mol=None,
set_mol_name=None,
read_model=None,
set_model_num=None,
verbosity=False)
Method for loading structures from a XYZ file. |
source code
|
|
|
|
rotate(self,
R=None,
origin=None,
model=None,
atom_id=None)
Rotate the structural information about the given origin. |
source code
|
|
|
|
translate(self,
T=None,
model=None,
atom_id=None)
Displace the structural information by the given translation vector. |
source code
|
|
|
|
|
|
|
write_pdb(self,
file,
model_num=None)
Method for the creation of a PDB file from the structural data. |
source code
|
|
|
Inherited from api_base.Base_struct_API:
__init__,
attached_atom,
empty,
from_xml,
get_model,
model_loop,
num_models,
num_molecules,
pack_structs,
target_mol_name,
to_xml,
validate
|