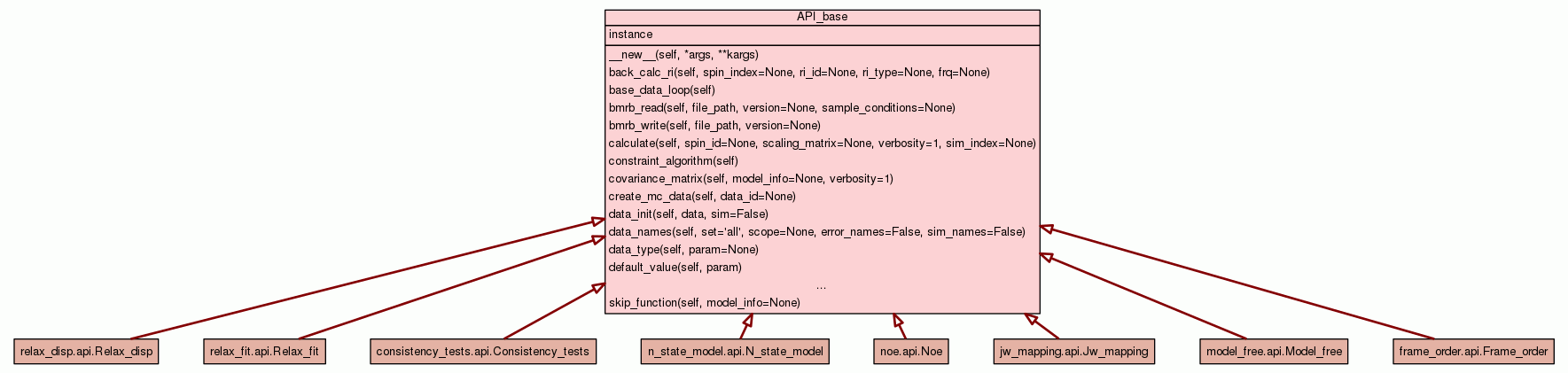
Base class defining the specific_analyses API.
All the methods here are prototype methods. To identify that the
method is not available for certain analysis types, if called a
RelaxImplementError is raised if called.
|
float
|
back_calc_ri(self,
spin_index=None,
ri_id=None,
ri_type=None,
frq=None)
Back-calculation of relaxation data. |
source code
|
|
|
anything
|
|
|
|
bmrb_read(self,
file_path,
version=None,
sample_conditions=None)
Prototype method for reading the data from a BMRB NMR-STAR formatted
file. |
source code
|
|
|
|
bmrb_write(self,
file_path,
version=None)
Prototype method for writing the data to a BMRB NMR-STAR formatted
file. |
source code
|
|
|
|
calculate(self,
spin_id=None,
scaling_matrix=None,
verbosity=1,
sim_index=None)
Calculate the chi-squared value. |
source code
|
|
|
str
|
|
|
numpy rank-2 array, numpy rank-2 array
|
covariance_matrix(self,
model_info=None,
verbosity=1)
Return the Jacobian and weights required for parameter errors via the
covariance matrix. |
source code
|
|
|
list of floats
|
|
|
|
|
|
list of str
|
data_names(self,
set='all',
scope=None,
error_names=False,
sim_names=False)
Return a list of names of data structures. |
source code
|
|
|
any type
|
|
|
float
|
|
|
|
|
|
|
duplicate_data(self,
pipe_from=None,
pipe_to=None,
model_info=None,
global_stats=False,
verbose=True)
Duplicate the data specific to a single model. |
source code
|
|
|
bool
|
|
|
class instance
|
|
|
list of str
|
|
|
list of str
|
|
|
|
grid_search(self,
lower=None,
upper=None,
inc=None,
scaling_matrix=None,
constraints=True,
verbosity=1,
sim_index=None)
Grid search method. |
source code
|
|
|
bool
|
|
|
bool
|
|
|
list of float
|
|
|
|
minimise(self,
min_algor=None,
min_options=None,
func_tol=None,
grad_tol=None,
max_iterations=None,
constraints=False,
scaling_matrix=None,
verbosity=0,
sim_index=None,
lower=None,
upper=None,
inc=None)
Minimisation method. |
source code
|
|
|
str
|
|
|
anything
|
|
|
tuple of (int, int, float)
|
|
|
str
|
|
|
|
molmol_macro(self,
data_type,
style=None,
colour_start=None,
colour_end=None,
colour_list=None,
spin_id=None)
Create and return an array of Molmol macros. |
source code
|
|
|
int
|
|
|
|
|
|
|
|
|
|
pymol_macro(self,
data_type,
style=None,
colour_start=None,
colour_end=None,
colour_list=None,
spin_id=None)
Create and return an array of PyMOL macros. |
source code
|
|
|
float
|
|
|
list of float
|
|
|
str or None
|
|
|
list of float
|
return_error(self,
data_id=None)
Return the error points corresponding to the data points used in
optimisation. |
source code
|
|
|
list of float
|
return_error_red_chi2(self,
data_id=None)
Return the error points corresponding to the overall gauss
distribution described by the STD_fit of the goodness of fit, where
STD_fit = sqrt(chi2/(N-p)). |
source code
|
|
|
str
|
|
|
str
|
|
|
str
|
|
|
tuple of length 2 of floats or None
|
return_value(self,
spin,
param,
sim=None,
bc=False)
Return the value and error corresponding to the parameter. |
source code
|
|
|
|
|
|
|
set_param_values(self,
param=None,
value=None,
index=None,
spin_id=None,
error=False,
force=True)
Set the model parameter values. |
source code
|
|
|
|
|
|
|
|
|
|
sim_init_values(self)
Initialise the Monte Carlo parameter values. |
source code
|
|
|
|
|
|
list of float or float
|
|
|
list of float
|
|
|
list of int
|
|
|
bool
|
|
|
Inherited from object:
__delattr__,
__format__,
__getattribute__,
__hash__,
__init__,
__reduce__,
__reduce_ex__,
__repr__,
__setattr__,
__sizeof__,
__str__,
__subclasshook__
|