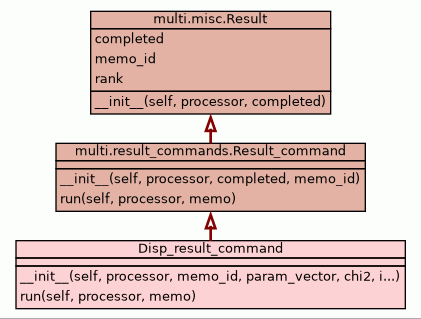
__init__(self,
processor=None,
memo_id=None,
param_vector=None,
chi2=None,
iter_count=None,
f_count=None,
g_count=None,
h_count=None,
warning=None,
missing=None,
back_calc=None,
completed=True)
(Constructor)
| source code
|
Set up this class object on the slave, placing the minimisation
results here.
- Parameters:
processor (multi.processor.Processor instance) - The processor object.memo_id (str) - The memo identification string.param_vector (numpy rank-1 array) - The optimised parameter vector.chi2 (float) - The final target function value.iter_count (int) - The number of optimisation iterations.f_count (int) - The total function call count.g_count (int) - The total gradient call count.h_count (int) - The total Hessian call count.warning (str or None) - Any optimisation warnings.missing (numpy rank-3 array) - The data structure indicating which R2eff/R1rho' base data is
missing.back_calc (numpy rank-3 array) - The back-calculated R2eff/R1rho' data structure from the target
function class. This is will be transfered to the master to be
stored in the r2eff_bc data structure.completed (bool) - A flag which if True signals that the optimisation successfully
completed. - Overrides:
multi.misc.Result.__init__
|