Relaxation dispersion analysis
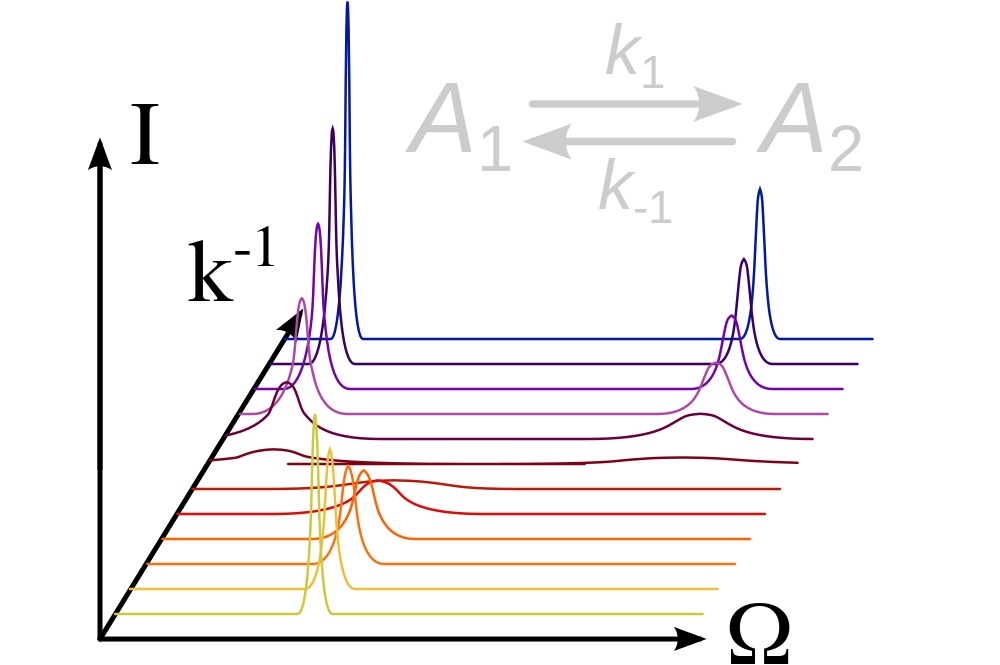
The software relax supports the analysis of NMR relaxation dispersion data (see Morin et al., 2014). All dispersion data types are supported as well as an almost complete list of relaxation dispersion models, both analytic and numeric.
Data types
The following relaxation dispersion data types are supported by relax:
- Single quantum (SQ) CPMG-type data.
- Zero quantum (ZQ) CPMG-type data.
- Double quantum (DQ) CPMG-type data.
- Multiple quantum (MQ) CPMG-type data.
- Combined proton-heteronuclear SQ, ZQ, DQ and MQ CPMG-type data. This is labelled as multiple MQ or MMQ data.
- R1ρ-type data.
Models
Numerous analytic and numeric relaxation dispersion models are supported by relax. It the model you are interested is not yet supported, please see the tutorial for adding dispersion models to relax.
Experiment independent models
The following models are independent of the experiment type:
- 'R2eff': This is the model used to determine the R2eff or R1ρ values and errors required as the base data for all other models. (See the wiki, manual, API documentation (for exponential curves), API documentation (for the 2-point fit)).
- 'No Rex': This is the model for no chemical exchange being present. (See the wiki, manual, API documentation).
Analytic models for SQ CPMG-type experiments
The following analytic models are designed for the standard single quantum CPMG-type experiments:
- 'LM63': The original Luz and Meiboom 1963 2-site fast exchange equation with parameters {R20, ..., Φex, kex}. (See the wiki, manual, API documentation).
- 'LM63 3-site': The original Luz and Meiboom 1963 3-site fast exchange equation with parameters {R20, ..., ΦexB, kB, ΦexC, kC}. (See the wiki, manual, API documentation).
- 'CR72': The reduced Carver and Richards 1972 2-site equation for most time scales whereby the simplification R2A0 = R2B0 is assumed. It has the parameters {R20, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'CR72 full': The full Carver and Richards 1972 2-site equation for most time scales with the parameters {R2A0, R2B0, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'IT99': The Ishima and Torchia 1999 2-site model for all time scales with pA » pB and with parameters {R20, ..., pA, Δω, tex}. (See the wiki, manual, API documentation).
- 'TSMFK01': The Tollinger et al., 2001 2-site very-slow exchange model for time scales within range of microsecond to second time scale. Applicable in the limit of slow exchange, when R2A0 - R2B0 « kAB, kBA « 1/τCPMG. 2*τCPMG is the time between successive 180 degree pulses. The parameters are {R2A0, ..., Δω, kAB}. (See the wiki, manual, API documentation).
- 'B14': The reduced Baldwin 2014 2-site equation for all time scales whereby the simplification R2A0 = R2B0 is assumed. It has the parameters {R20, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'B14 full': The full Baldwin 2014 2-site equation for all time scales with the parameters {R2A0, R2B0, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
Numeric models for SQ CPMG-type experiments
The following analytic models are designed for the standard single quantum CPMG-type experiments:
- 'NS CPMG 2-site expanded': A model for 2-site exchange expanded using Maple by Nikolai Skrynnikov (Tollinger et al., 2001). It has the parameters {R20, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'NS CPMG 2-site 3D': The reduced model for 2-site exchange using 3D magnetisation vectors whereby the simplification R2A0 = R2B0 is assumed. It has the parameters {R20, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'NS CPMG 2-site 3D full': The reduced model for 2-site exchange using 3D magnetisation vectors with the parameters {R2A0, R2B0, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'NS CPMG 2-site star': The reduced model for 2-site exchange using complex conjugate matrices whereby the simplification R2A0 = R2B0 is assumed. It has the parameters {R20, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'NS CPMG 2-site star full': The full model for 2-site exchange using complex conjugate matrices with the parameters {R2A0, R2B0, ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
Analytic models for MMQ-type data
The following analytic models are designed for combined proton-heteronuclear SQ, ZQ, DQ and MQ (MMQ, or multiple MQ) CPMG-type experiments:
- 'MMQ CR72': The Carver and Richards (1972) 2-site model for most time scales expanded for MMQ CPMG data by Korzhnev et al., 2004. It has the parameters {R20, ..., pA, Δω, ΔωH, kex}. (See the wiki, manual, API documentation).
Numeric models for MMQ-type data
The following numeric models are designed for MMQ CPMG-type experiments:
- 'NS MMQ 2-site': The model for 2-site exchange whereby the simplification R2A0 = R2B0 is assumed. It has the parameters {R20, ..., pA, Δω, ΔωH, kex}. (See the wiki, manual, API documentation).
- 'NS MMQ 3-site linear': The model for 3-site exchange linearised with kAC = kCA = 0 whereby the simplification R2A0 = R2B0 = R2C0 is assumed. It has the parameters {R20, ..., pA, ΔωAB, ΔωBC, ΔωABH, ΔωBCH, kexAB, kexBC, kexAC}. (See the wiki, manual, API documentation).
- 'NS MMQ 3-site': The model for 3-site exchange whereby the simplification R2A0 = R2B0 = R2C0 is assumed. It has the parameters {R20, ..., pA, ΔωAB, ΔωBC, ΔωABH, ΔωBCH, kexAB, kexBC, kexAC}. (See the wiki, manual, API documentation).
Analytic models for R1rho experiments
The following analytic models are designed for R1ρ-type experiments:
- 'M61': The Meiboom 1961 2-site fast exchange equation for on-resonance data with parameters {R1ρ', ..., Φex, kex}. (See the wiki, manual, API documentation).
- 'M61 skew': The Meiboom 1961 2-site equation for all time scales with pA » pB and with parameters {R1ρ', ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'DPL94': The Davis et al., 1994 extension of the M61 model for off-resonance data with parameters {R1ρ', ..., Φex, kex}. (See the wiki, manual, API documentation).
- 'TP02': The Trott and Palmer 2002 2-site equation for all time scales with pA » pB and with parameters {R1ρ', ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'TAP03': The Trott et al., 2003 off-resonance 2-site analytic equation for all time scales with the weak condition pA » pB and with parameters {R1ρ', ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'MP05': The Miloushev and Palmer 2005 off-resonance 2-site equation for all time scales with parameters {R1ρ', ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
Numeric models for R1rho experiments
The following numeric models are designed for R1ρ-type experiments:
- 'NS R1rho 2-site': The model for 2-site exchange using 3D magnetisation vectors. It has the parameters {R1ρ', ..., pA, Δω, kex}. (See the wiki, manual, API documentation).
- 'NS R1rho 3-site linear': The model for 3-site exchange linearised with kAC = kCA = 0 whereby the simplification R1ρA' = R1ρB' = R1ρC' is assumed. It has the parameters {R1ρ', ..., pA, pB, ΔωAB, ΔωBC, kexAB, kexBC}. (See the wiki, manual, API documentation).
- 'NS R1rho 3-site': The model for 3-site exchange whereby the simplification R1ρA' = R1ρB' = R1ρC' is assumed. It has the parameters {R1ρ', ..., pA, pB, ΔωAB, ΔωBC, kexAB, kexBC, kexAC}. (See the wiki, manual, API documentation).
